Computing, Environment and Life Sciences
Advanced Protein Characterization Facility
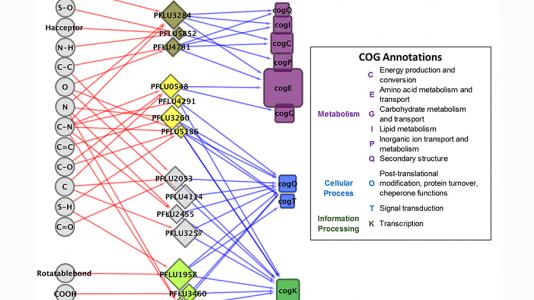
APCFThe Advanced Protein Characterization Facility (APCF) utilizes high-throughput automated methods for bioinformatics, proteomics, molecular and structural biology.
Proteins are the molecular machines of all cells, and as with any machine, it is impossible to understand how a protein works without knowing what it looks like — that is, imaging its three-dimensional structure. The Advanced Protein Characterization Facility (APCF) will help us to “see” proteins more quickly and with higher proficiency than before. The increase in space emboldens plans to pursue additional programmatic support and new equipment. The APCF also supports Argonne’s ongoing experimental and computational systems work in biology, funded by both the U.S. Department of Energy and the National Institutes of Health.